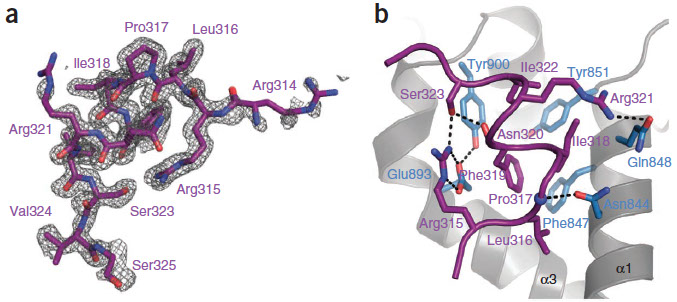


1) Human Argonaute 2 MID domain
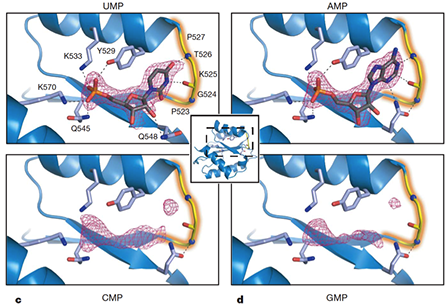
MicroRNAs (miRNAs) mediate post-transcriptional gene regulation through association with Argonaute proteins (AGOs). Crystal structures of archaeal and bacterial homologues of AGOs have shown that the MID(middle) domain mediates the interaction with the phosphorylated 5' end of the miRNA guide strand and this interaction is thought to be independent of the identity of the 5' nucleotide in these systems. However, analysis of the known sequences of eukaryotic miRNAs and co-immunoprecipitation experiments indicate that there is a clear bias for U or A at the 5' position. Here we report the crystal structure of a MID domain from a eukaryotic AGO protein, human AGO2. The structure, in complex with nucleoside monophosphates (AMP, CMP, GMP, and UMP) mimicking the 5' end of miRNAs, shows that there are specific contacts made between the base of UMP or AMP and a rigid loop in the MID domain. Notably, the structure of the loop discriminates against CMP and GMP and dissociation constants calculated from NMR titration experiments confirm these results, showing that AMP (0.26 mM) and UMP (0.12mM) bind with up to 30-fold higher affinity than either CMP (3.6mM) or GMP (3.3mM). This study provides structural evidence for nucleotide specific interactions in theMID domain of eukaryotic AGO proteins and explains the observed preference for U or A at the 5' end of miRNAs.
Click to see a movie of the hAGO structure
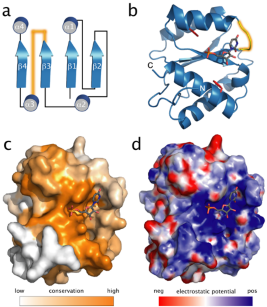
2) Arabidopsis Argonaute MID domains
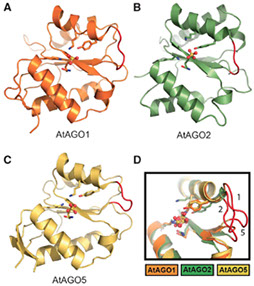
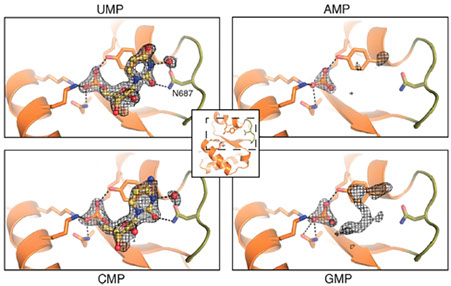
The 5'-nucleotide of small RNAs associates directly with the MID domain of Argonaute (AGO) proteins. In humans, the identity of the 5'-base is sensed by the MID domain nucleotide specificity loop and regulates the integrity of miRNAs. In Arabidopsis thaliana, the 5'-nucleotide also controls sorting of small RNAs into the appropriate member of the AGO family; however, the structural basis for this mechanism is unknown. Here, we present crystal structures of the MID domain from three Arabidopsis AGOs, AtAGO1, AtAGO2 and AtAGO5, and characterize their interactions with nucleoside monophosphates (NMPs). In AtAGOs, the nucleotide specificity loop also senses the identity of the 5'-nucleotide but uses more diverse modes of recognition owing to the greater complexity of small RNAs found in plants. Binding analyses of these interactions reveal a strong correlation between their affinities and evolutionary conservation.
2) CCR4-NOT1 fragment
Tristetraprolin (TTP) is an RNA-binding protein that controls the inflammatory response by limiting the expression of several proinflammatory cytokines. TTP post-transcriptionally represses gene expression by interacting with AU-rich elements (AREs) in 3′ untranslated regions of target mRNAs and subsequently engenders their deadenylation and decay. TTP accomplishes these tasks, at least in part, by recruiting the multisubunit CCR4–NOT deadenylase complex to the mRNA. Here we identify an evolutionarily conserved C-terminal motif in human TTP that directly binds a central domain of CNOT1, a core subunit of the CCR4–NOT complex. A high-resolution crystal structure of the TTP–CNOT1 complex was determined, providing the first structural insight, to our knowledge, into an ARE-binding protein bound to the CCR4–NOT complex. Mutations at the CNOT1-TTP interface impair TTP-mediated deadenylation, demonstrating the significance of this interaction in TTP-mediated gene silencing.